Stony Brook University has been at the forefront of developing innovative self supervised learning and diffusion methods to create Pathology foundation models for classification, segmentation and prediction tasks. In addition to our AI analysis pipelines and models, virtually all of which are publicly distributed, the group has created and distributed a variety of impactful tools.
Foundation Models and Self-Supervised Training
Visual homogeneity in many tissue regions can reduce effectiveness of self supervised training methods in Pathology. Pairwise self-supervised learning (SSL) methods like DINO (DIstillation with NO labels) operate on a simple yet effective principle, leveraging the power of self-distillation and self-supervised learning. The networks learn to identify and represent salient features of the data. This allows identification of visual elements without explicit labeling. DINO applies different augmentations to an input image and then passes these augmented versions through student and the teacher networks. The goal is for the student network to predict the same output as the teacher network, even though they see different versions of the same image.
Comparing attention maps arising from “natural” images such as a bird in a tree with Pathology images illustrates challenges arising from visual homogeneity seen in Pathology images. In (Figure 1) we see that attention maps concentrate on only a limited set of regions. While such sparsity might be advantageous in natural imaging tasks like object classification, it proves detrimental to computational pathology.
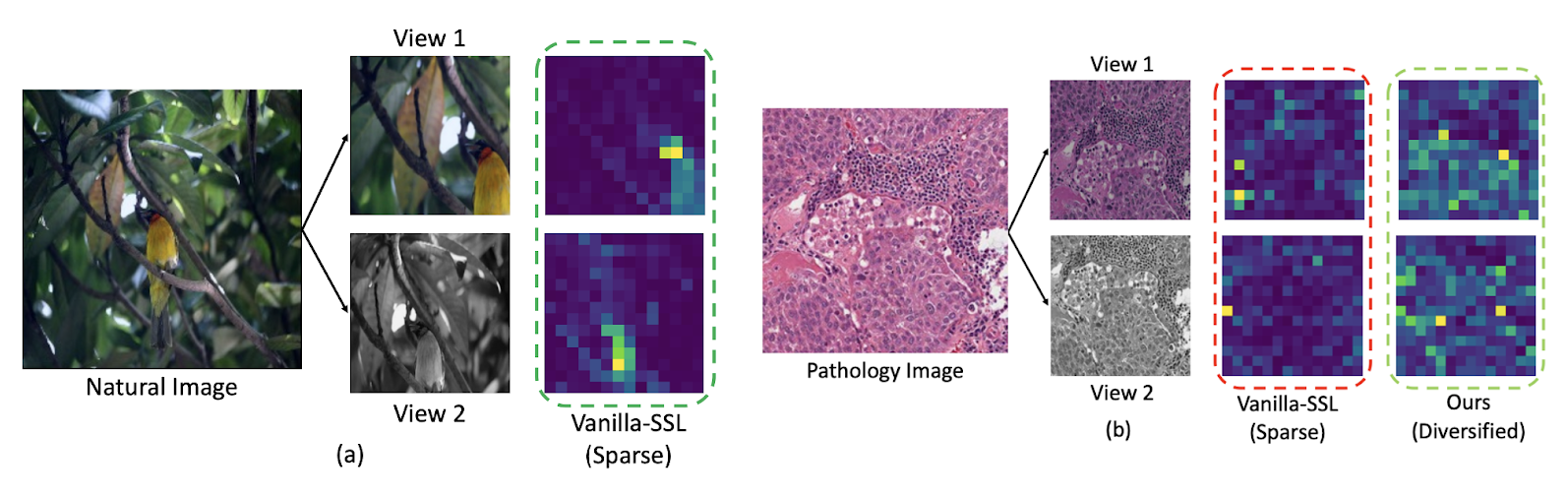
Figure 1 - Comparison of attention maps associated with “natural” images with attention maps generated from Pathology images. The attention map associated with the “vanilla” self supervised learning algorithm is sparse. This corresponds well to excellent performance on a task such as finding or classifying a bird (left panel) but a sparse attention map is often associated with poor performance in Pathology images (right panel)
The Digital Pathology team at SBU has developed self-supervised (SSL) training methods that are well adapted to Pathology images. Diversity-inducing Representation Learning (DiRL) is one example where we use supervised labeling to generate input for a pairwise self supervised training method. DiRL extracts and separately processes region-level representations for cellular and non-cellular regions in WSI crops, capturing the critical interactions between these regions—a vital aspect in digital pathology. We use cell segmentation/classification methods to identify regions with distinct cell populations and types; we then go on to use these labeled regions in our self supervised training methods. We demonstrate the efficacy of DiRL through slide-level and patch-level classification tasks. Results of this work were recently published in Medical Image Analysis
A complementary approach is to improve integration of spatial location into the matching methods used in self-supervised training. This method, published in Information Processing in Medical Imaging (IPMI) 2023 , is called precise location matching - see Figure 2 for an illustration. This method matches the representation of a local patch in a view to multiple corresponding overlapping patches in another. Our extensive experiments demonstrated the efficacy of this approach in semantic and instance segmentation tasks, significantly outperforming previous methods in terms of average precision for detection and instance segmentation.
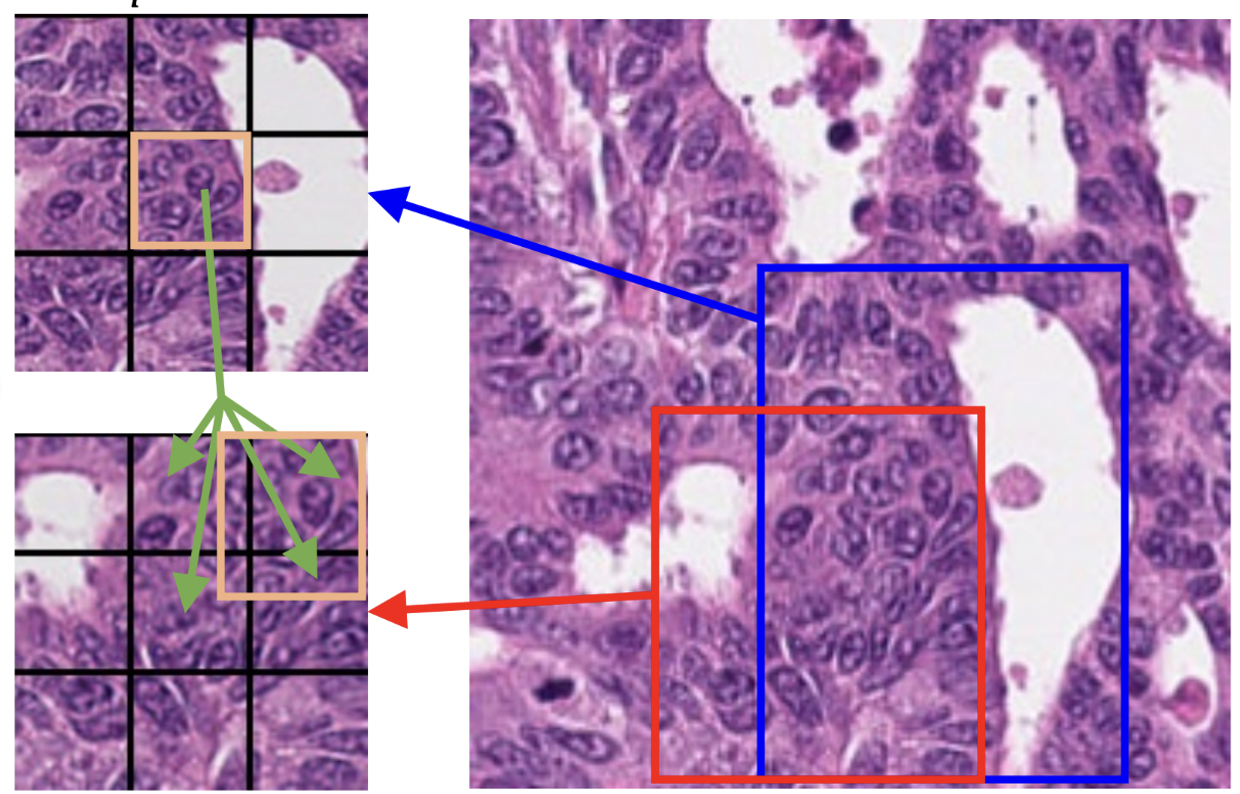
Figure 2: Precise Location Matching, spatial location information is incorporated when matching cropped/resized/transformed images
We are also developing innovative methods of adapting pre-trained general purpose foundation models for Pathology. SAM-Path published in a MICCAI 2023 workshop , developed by our group, starts with the Segment Anything Model (SAM), a foundation model developed by Meta’s FAIR laboratory was trained with over one billion segmentation masks. SAM-Path adapts the foundation model by introducing a pathology encoder component for semantic segmentation in digital pathology images. SAM-Path uses a novel trainable prompt approach, published in MICCAI 2023 which makes it possible for SAM to perform multi-class semantic segmentation. Additionally, we incorporate a pathology foundation model as an extra pathology encoder, providing domain-specific information. Our MICCAI 2023 paper demonstrates substantial improvement over the original SAM model. These contributions enhance SAM's capabilities and showcase its effectiveness in pathology image analysis.
Generative Models in Pathology
Our group is developing a variety of generative AI methods. To achieve high-quality results, diffusion models must be trained on large datasets. This can be notably prohibitive for models in specialized domains, such as computational pathology. Conditioning on labeled data is known to help in data-efficient model training. Therefore, histopathology reports, which are rich in valuable clinical information, are an ideal choice as guidance for a histopathology generative model. One approach is to condition on Pathology reports along with spatially mapped labels representing presence of tumors and of tumor infiltrating lymphocytes (TILs). This approach fuses image, Pathology report and tumor/TIL label information to generate highly realistic images as shown in Figure 3. A paper describing this work has been published in WACV 2024. In earlier work, we integrated ViT with diffusion autoencoders, that work was published at the DGM4 MICCAI 2023 workshop. In a complementary approach, we incorporate tools for spatial statistics and topological data analysis into deep generative models as both conditional inputs and a differentiable loss. This way, we are able to generate high quality multi-class cell layouts for the first time. We show that the topology-rich cell layouts can be used for data augmentation and improve the performance of downstream tasks such as cell classification. This work was published in CVPR 2023 .
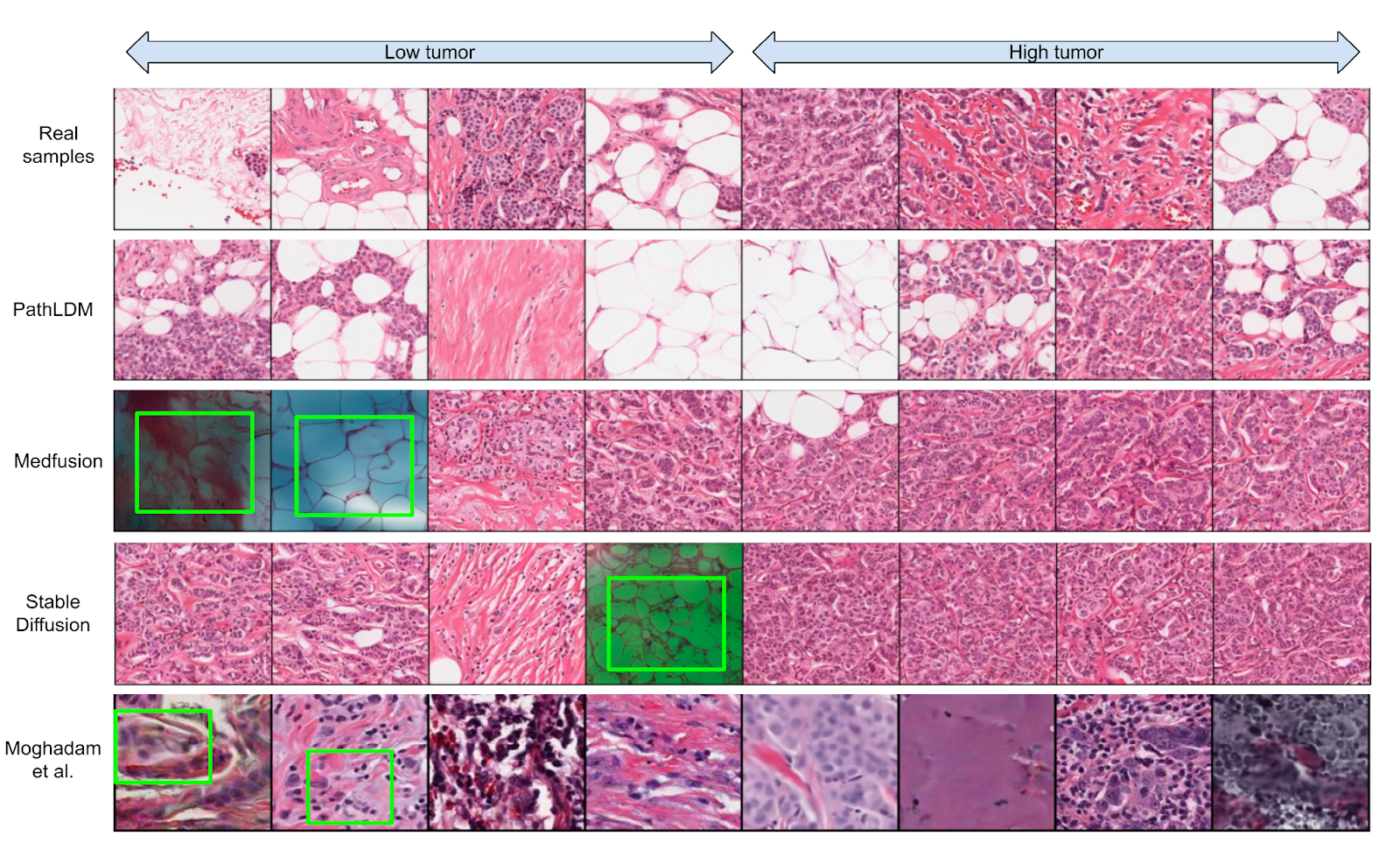
Figure 3. We choose a single text report and produce synthetic samples using Medfusion, Stable Diffusion, and PathLDM. Samples generated by Medfusion and Stable Diffusion show artifacts (indicated by green boxes) that are not present in our outputs. Moghadam et al. produces images in lower resolution (128 × 128) and exhibits artifacts and blurriness (green boxes in the last row) in the output images. The first row contains the real samples from the corresponding WSI.
Open Source Tools
WSInfer is an open-source software suite designed to make deep learning for pathology more streamlined and accessible. WSInfer applies trained patch classification methods to whole slide images either through a command line tool and Python package or through the widely adopted QuPath software platform. The offering includes a model zoo, which enables pathology models and metadata to be easily shared in a standardized form. Users can choose a model from the Zoo (offerings currently include tumor/TIL patch classification methods) or provide a local trained model along with a configuration file. This software was created in joint effort between the Stony Brook and University of Edinburgh QuPath group; description and links to software in NPJ Precision Oncology
Champkit is a Python-based software package that enables rapid exploration and evaluation of deep learning models for patch-level classification of histopathology data. ChampKit is designed to be highly reproducible and enables systematic, unbiased evaluation of patch-level histopathology classification. It incorporates public datasets for six clinically important tasks and access to hundreds of (pre-trained) deep learning models. It can easily be extended to custom patch classification datasets and custom deep learning architectures. This is published in Computer Methods and Programs in Biomedicine .
SBU Digital Pathology group and external Collaborators:
Faculty - Joel Saltz, Dimitris Samaras, Prateek Prasanna, Tahsin Kurc, Rajarsi Gupta, Chao Chen.
External Collaborators: Maria Vakalopoulou, Pete Bankhead, Alan O’Callaghan, Fiona Inglis, Swarad Gat, Srijan Das, Peter Koo and Beatrice Knudson. Graduate students: Saarthak Kapse, Jingwei Zhang, Srikar Yellapragada, Erich Bremer, Alexandros Graikos, Xuan Xu, Jakub Kaczmarzyk and Shahira Abousamra, Research Scientist Ke Ma

