Active Projects
QUIP
Integrating Images, Data and Knowledge in New Ways to Benefit Patients
 Dr. Tahsin Kurc explains:
Dr. Tahsin Kurc explains:
"It is highly desirable in research studies to use large datasets in order to obtain robust, statistically significant results, but the scale of an image-based study is often limited by how efficiently image datasets can be processed through image segmentation, feature computation, and classification pipelines. Modern HPC systems provide significant processing power, through clusters of hybrid computation nodes with multi-core CPUs and multiple graphics processing units (GPUs), and memory capacity, distributed across computation nodes or accessible via shared-memory mechanisms. Nevertheless, implementing analysis applications on HPC systems is not an easy task, because of the heterogeneous nature, complexity, and scale of the contemporary systems. Our work researches and develops methods and runtime middleware systems that can carry out high throughput processing of large numbers of images by coordinated use of multi-core CPUs and GPUs on computing clusters.
"Algorithm evaluation provides a means to characterize variability across image analysis algorithms, validate algorithms by comparison of multiple results, and facilitate algorithm sensitivity studies. Sensitivity quantification process in algorithm evaluation is a data intensive and computationally expensive process that involves processing datasets using variations of an analysis pipeline, comparing results from these variations, and quantifying agreements and disagreements between the results. Analysis parameter tuning is another important process in which the parameter space of an analysis workflow is searched by comparing analysis results with ground truth to find the set of parameters, which produces results that are closest to the ground truth with respect to some comparison metric. The sizes of images and analysis results in pathology image analysis pose significant challenges in these processes. We develop methods and an integrated software framework that addresses the processing and data management challenges of sensitivity quantification and parameter tuning by carefully distributing and coordinating operations and data across multiple machines, multi-core CPUs and co-processors and by reducing data movement and computation costs.
"Understanding complex mechanisms underlying a disease requires integration and mining of information obtained at multiple scales and across multiple data types. Our work develops techniques and data structures for multi-resolution and dynamic representations of very large volumes of primary and derived data. These techniques are supported by tools that allow a scientist to carry out correlative and exploratory queries on data."
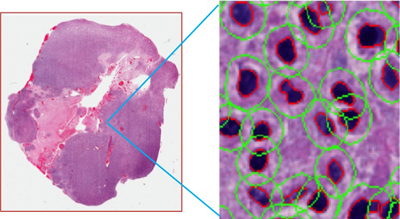
Mug Shots of Biological Bad Guys
A primary research focus in BMI is the development of frameworks, techniques, information models, and high performance computing support for the integrative analysis of high resolution microscopy images obtained from tissues. This research is driven by the vision of delivering the next generation of integrative information technologies, which facilitate truly quantitative data-directed healthcare. These technologies will make it possible to efficiently:
- interrogate and explore data at multiple scales and dimensions
- integrate it across scales and dimensions, and
- extract and compare information across individuals and cohorts.
Clinicians and researchers would be able to study common and discriminating biological characteristics across cohorts of subjects, to categorize patient subpopulations and determine targeted prevention and treatment strategies tailored to individual patients.
People
Joel Saltz, Tahsin Kurc, Allen Tannenbaum, Margaret Schedel, James Davis, Arie Kaufman and Donald Harrington
Past Projects
Informatics for Integrative Brain Tumor Whole Slide Analysis - funded by the National Library of Medicine, this project develops, deploys, and evaluates methodologies, information models, tools, and analytic pipelines that will make it feasible to systematically carry out large-scale comparative analyses of brain tumor histological features using whole slide images and of patterns of protein and gene expression. The research and development effort involves (1) highly optimized algorithms and analytic pipelines which enable investigators to carry out large-scale comparative analyses of brain tumor histological features using whole slide images and of patterns of protein and gene expression, (2) flexible information models to manage information associated with analysis of brain tumor whole virtual slide data, (3) runtime systems that take advantage of high performance computing platforms to scale image analyses to large datasets. The methods and tools will be used to determine the relationship between image-based tumor signatures and clinical outcome, gene expression category, genetic gains/losses and methylation status and map the activity of signal transduction pathways and transcriptional networks relative to the tumor microenvironment using quantitative multiplex quantum dot immunohistochemistry and histology feature descriptions. This project is a collaborative effort between Stony Brook University (Joel Saltz, Tahsin Kurc), Emory University (Daniel Brat, Lee Cooper, David Gutman, Fusheng Wang, Jun Kong, Roberd Bostick, Carlos Moreno), and Rutgers Cancer Institute of New Jersey (David J. Foran).
In Silico Brain Tumor Research Center - funded by the National Cancer Institute as one of the In Silico Research Centers of Excellence, this center explores new ideas in brain tumor translational research through multi-scale, integrative in silico experiments. The experiments make use of molecular data, pathology image data, radiology data, and clinical outcome data. The experiments involve execution of pipelines of image processing operations on large image datasets, execution of multiple bioinformatics analysis methods, and comparison and correlation of data from multiple data types and sources. The center is a collaborative effort between Stony Brook University, Emory University, Henry Ford Hospital, and Thomas Jefferson University.
Image Mining for Comparative Analysis of Expression Patterns in Tissue Microarray - funded by the National Institutes of Health, this project designs, implements, and evaluates (1) a new family of multi-stage, searching algorithms to facilitate quick, reliable interrogation of large-scale, clinical and research, microscopy applications including whole-slide imaging and tissue microarray; (2) high-throughput services capable of automatically detecting, archiving and indexing user-specified objects (e.g. tissues, cells) in large collections of images and implement extensions to the data models and support for optimized pipeline selection, (3) optimized imaging, computational and content-based image retrieval algorithms and tools using a wide range of different tissues, cancer types and biomarkers to support clinical and research experiments and studies involving patient stratification, quality-control, and outcomes assessment. These capabilities will be deployed as analytical tools, data models, user-centered interfaces and reference libraries of imaged specimens to make them available to the clinical and research communities to support future development and testing of new hypotheses, algorithms and methods. This is a collaborative project between Rutgers Cancer Institute of New Jersey (David J. Foran), Stony Brook University (Joel Saltz, Tahsin Kurc), and University of Kentucky (Lin Yang).
Related Publications
- G. Teodoro, T. Pan, T. Kurc, J. Kong, L. Cooper, and J. Saltz: Efficient Irregular Wavefront Propagation Algorithms on Hybrid CPU-GPU Machines, Parallel Computing, 39(4-5), 189-211, 2013. [paper]
- J. Saltz, G. Teodoro, T. Pan, L. Cooper, J. Kong, S. Klasky, T. Kurc: Feature-based analysis of large-scale spatio-temporal sensor data on hybrid architectures, International Journal of High Performance Computing Applications, 27(3), pp. 263-272, 2013. [paper]
- F. Wang, J. Kong, J. Gao, L. Cooper, T. Kurc, Z. Zhou, D. Adler, C. Vergara-Niedermayr, B. Katigbak, D. Brat, J. Saltz: A high-performance spatial database based approach for pathology imaging algorithm evaluation, Journal of pathology informatics, 4, 2013. [paper]
- L.A. Cooper, J. Kong, D.A. Gutman, F. Wang, J. Gao, C. Appin, S. Cholleti, T. Pan, A. Sharma, L. Scarpace, T. Mikkelsen, T. Kurc, C.S. Moreno, D.J. Brat, J.H. Saltz: Integrated morphologic analysis for the identification and characterization of disease subtypes. J Am Med Inform Assoc 19 (2012) 317-323. [paper]
- L.A. Cooper, D.A. Gutman, C. Chisolm, C. Appin, J. Kong, Y. Rong, T. Kurc, E.G. Van Meir, J.H. Saltz, C.S. Moreno, D.J. Brat: The Tumor Microenvironment Strongly Impacts Master Transcriptional Regulators and Gene Expression Class of Glioblastoma. Am J Pathol, 2012. [paper]
- L. Cooper, A. Carter, A. Farris, F. Wang, J. Kong, D. Gutman, P. Widener, T. Pan, S. Cholleti, A. Sharma, T. Kurc, D. Brat, J. Saltz: Digital Pathology: Data Intensive Frontier in Medical Imaging. IEEE Special Bicentennial Issue on Audiovisual Frontiers, 100(4), pp. 991-1003, 2012. [paper]
- J. Kong, L.A. Cooper, F. Wang, D.A. Gutman, J. Gao, C. Chisolm, A. Sharma, T. Pan, E.G. Van Meir, T. Kurc, C.S. Moreno, J.H. Saltz, D.J. Brat: Integrative, multimodal analysis of glioblastoma using TCGA molecular data, pathology images, and clinical outcomes. IEEE Trans Biomed Eng 58 (2011) 3469-3474. [paper]
- F. Wang, J. Kong, L. Cooper, T. Pan, T. Kurc, W. Chen, A. Sharma, C. Niedermayr, T.W. Oh, D. Brat, A.B. Farris, D.J. Foran, J. Saltz: A data model and database for high-resolution pathology analytical image informatics. J Pathol Inform 2 (2011) 32.[paper]
- D Foran, L Yang, W Chen, J Hu, L Goodell, M Reiss, F Wang, T Kurc, T Pan, A Sharma, J Saltz. ImageMiner: A Software System for Comparative Analysis of Tissue Microarrays Using Content-Based Image Retrieval, High-Performance Computing, and Grid Technology. Journal of the American Medical Informatics Association. May 23, 2011. 18:352-353. PMID: 21606133. [paper]
- L.A. Cooper, J. Kong, D.A. Gutman, F. Wang, S.R. Cholleti, T.C. Pan, P.M. Widener, A. Sharma, T. Mikkelsen, A.E. Flanders, D.L. Rubin, E.G. Van Meir, T. Kurc, C.S. Moreno, D.J. Brat, J.H. Saltz: An integrative approach for in silico glioma research. IEEE Trans Biomed Eng 57 (2010) 2617-2621. [paper]
- L.A. Cooper, D.A. Gutman, Q. Long, B.A. Johnson, S.R. Cholleti, T. Kurc, J.H. Saltz, D.J. Brat, C.S. Moreno: The proneural molecular signature is enriched in oligodendrogliomas and predicts improved survival among diffuse gliomas. PLoS ONE 5 (2010) e12548. [paper]
- V.S. Kumar, T. Kurc, V. Ratnakar, J. Kim, G. Mehta, K. Vahi, Y.L. Nelson, P. Sadayappan, E. Deelman, Y. Gil, M. Hall and J. Saltz: Parameterized Specification, Configuration and Execution of Data-Intensive Scientific Workflows. Cluster Computing: the Journal of Networks, Software Tools and Applications, Special Issue on High Performance Distributed Computing, Vol. 13(3), pp. 315-333, 2010. [paper]
- A. Sharma, T. Pan, B. Cambazoglu, M. Gurcan, T. Kurc, J. Saltz: VirtualPACS-A Federating Gateway to Access Remote Image Data Resources over the Grid. J. Digital Imaging, Vol. 22(1), pp. 1-10, 2009. [paper]
- V. S. Kumar, S. Narayanan, T. Kurc, J. Kong, M. N. Gurcan, J. H. Saltz, ”Analysis and Semantic Querying in Large Biomedical Image Datasets”, IEEE Computer Magazine, special issue on Data-Intensive Computing, Vol. 41(4), pp. 52-59, 2008. [paper]
- V. S. Kumar, B. Rutt, T. Kurc, U. V. Catalyurek, T. C. Pan, S. Chow, S. Lamont, M. Martone, J. H. Saltz, ”Large-scale Biomedical Image Analysis in Grid Environments”, IEEE Transactions on Information Technology in Biomedicine, Vol. 12(2), pp. 154-161, 2008. [paper]
- K. Mosaliganti, T. C. Pan, R. Ridgway, R. Sharp, L. Cooper, A. Gulacy, A. Sharma, O. Irfanoglu, R. Machiraju, T. Kurc, P. Wenzel, A. de Bruin, G. Leone, J. H. Saltz, K. Huang, ”An Imaging Workflow for Characterizing Phenotypical Change in Large Histological Mouse Model Datasets”, Journal of Biomedical Informatics, Vol. 41(6), pp. 863-873, 2008. [paper]
- T. Pan, M. Gurcan, S. Langella, S. Oster, S. Hastings, A. Sharma, B. Rutt, D. Ervin, T. Kurc, K. Siddiqui, J. Saltz, E. Siegel, ”GridCAD: A Grid-Based Computer-Aided Detection System”, Radio-graphics, Vol. 27, pp. 889-897, 2007. [paper]
- M. Gurcan, T. Pan, A. Sharma, T. Kurc, S. Oster, S. Langella, S. Hastings, K. Siddiqui, E. Siegel, J. Saltz: GridImage: A Novel Use of Grid Computing to Support Interactive Human and Computer-Assisted Detection Decision Support. Journal of Digital Imaging, Vol. 20(2), pp. 160-171, 2007. [paper]
- G. Teodoro, T. Pan, T. Kurc, J. Kong, L. A. Cooper, N. Podhorszki, S. Klasky, J. Saltz, "High-throughput Analysis of Large Microscopy Image Datasets on CPU-GPU Cluster Platforms," in the 27th IEEE International Parallel and Distributed Processing Symposium (IPDPS), Boston, Massachusetts, USA. May 20-24, 2013. [paper]
- P. Widener, T. Kurc, W. Chen, F. Wang, L. Yang, J. Hu, V. Kumar, V. Chu, L. Cooper, J. Kong, A. Sharma, T. Pan, J. Saltz, and D. Foran: High Performance Computing Techniques for Scaling Image Analysis Workflows. Lecture Notes in Computer Science, Applied Parallel and Scientific Computing (PARA 10), Springer, p67-77, 2012. [paper]
- F. Wang, T. W. Oh, C. Vergara-Niedermayr, T. Kurc, and J. Saltz: Managing and Querying Whole Slide Images. Proceedings of SPIE Medical Imaging, Feb 4-9, 2012. San Diego, California, USA. [paper]
- G. Teodoro, T. Kurc, T. Pan, L. Cooper, J. Kong, P. Widener, and J. Saltz: Accelerating Large Scale Image Analyses on Parallel, CPU-GPU Equipped Systems. The 26th IEEE International Parallel and Distributed Processing Symposium (IPDPS), May 2012. [paper]
- J. Kong, L. Cooper, F. Wang, C. Chisolm, C. Moreno, T. Kurc, P. Widener, D. Brat, J. Saltz: A Comprehensive Framework for Classification of Nuclei in Digital Microscopy Imaging: An Application to Diffuse Gliomas. Proc IEEE Int Symp Biomed Imaging (2011) 2128-2131. [paper]
- L.A. Cooper, J. Kong, F. Wang, T. Kurc, C.S. Moreno, D.J. Brat, J.H. Saltz: Morphological Signatures and Genomic Correlates in Glioblastoma. Proc IEEE Int Symp Biomed Imaging (2011) 1624-1627. [paper]
- Jun Kong, Lee Cooper, T. Kurc, Daniel Brat, and Joel Saltz: Towards Building Computerized Image Analysis Framework for Nucleus Discrimination in Microscopy Images of Diffuse Glioma. The 33rd International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 6605-6608, Boston, MA, August, 2011. [paper]
- P. Widener, T. Kurc, W. Chen, F. Wang, L. Yang, J. Hu, V. Kumar, V. Chu, L. Cooper, J. Kong, A. Sharma, T. Pan, J. Saltz, D. Foran: Grid-Enabled, High-performance Microscopy Image Analysis. The 2nd International Workshop on High-Performance Medical Image Computing for Image-Assisted Clinical Intervention and Decision-Making (HP-MICCAI 2010) Beijing, China, September 2010.
- F. Wang, T. Kurc, P. Widener, T. Pan, J. Kong, L. Cooper, D. Gutman, A. Sharma, S. Cholleti, V. Kumar and J. Saltz: High-performance Systems for In Silico Microscopy Imaging Studies. The 7th International Conference on Data Integration in the Life Sciences, Gothenburg, Sweden, August 2010. [paper]
- J. Saltz, T. Kurc, S. Cholleti, J. Kong, C. Moreno, A. Sharma, T. Pan, L. Cooper, D. Gutman, E. Van Meir, T. Mikkelsen, A. Flanders, D. Rubin, D. Brat, Multi-Scale, Integrative Study of Brain Tumor: In Silico Brain Tumor Research Center, Proceedings of the Annual Symposium of American Medical Informatics Association 2010 Summit on Translational Bioinformatics (AMIA-TBI 2010), San Francisco, LA, March 2010. [paper]
- J. Kong, L. Cooper, A. Sharma, T. Kurc, D. Brat and J. Saltz: Texture Based Image Recognition in Microscopy Images of Diffuse Gliomas with Multi-class Gentle Boosting Mechanism. The 35th International Conference on Acoustics, Speech, and Signal Processing (ICASSP), pp.457-460, Dallas, TX, March 2010. [paper]
- V.S. Kumar, T. Kurc, G. Mehta, K. Vahi, V. Ratnakar, J. Kim, E. Deelman, Y. Gil, P. Sadayappan, M. Hall and J. Saltz, ”An Integrated Framework for Parameter-based Optimization of Scientific Workflows”, Proceedings of the ACM International Symposium on High Performance Distributed Computing (HPDC), June 2009. [paper]
- V. Kumar, T. Kurc, J. Kong, U. Catalyurek, M. Gurcan, J. Saltz, Performance vs. Accuracy Trade-offs for Large-scale Image Analysis Applications, Cluster 2007, 2007. [paper]

